Developing Therapies to Combat Drug-Resistant Cell States
Single-cell proteomics can predict resistance and enable therapies to combat drug-resistant cell states
Drug resistant tumors present challenges in cancer treatment
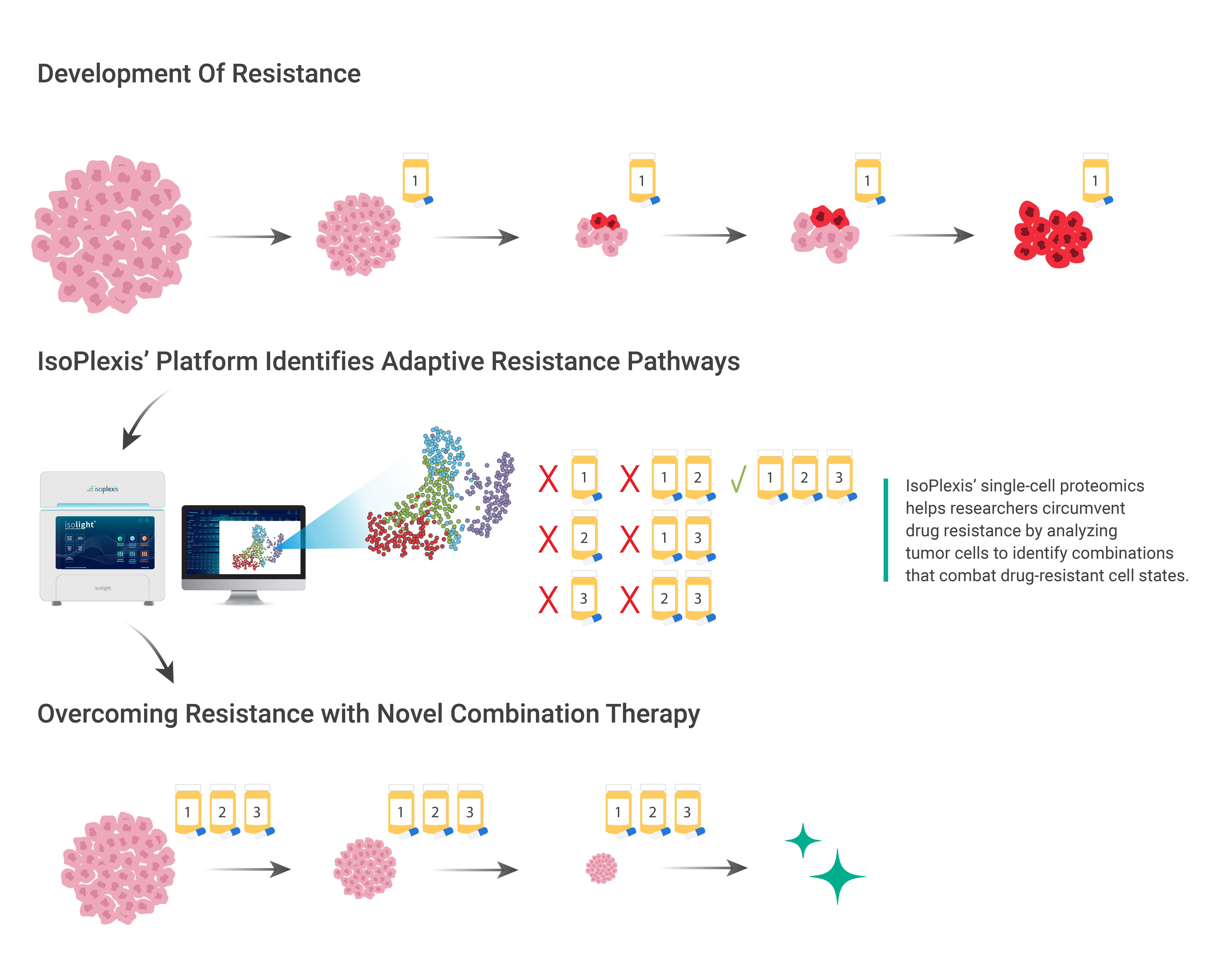
A significant challenge in the treatment of cancers is the ability of some cancers to develop drug resistance, leading to poor response to treatment or recurrence after treatment. However, cutting-edge technologies now allow researchers to characterize the mechanisms of drug resistance, enabling the development of individualized combination therapies to circumvent patients’ resistance.
IsoPlexis’ unique functional proteomics platform is the emerging standard in single-cell and ultra-low volume sample proteomics. With its multi-omic approach facilitating analysis of intracellular proteins, this technology is accelerating the discovery of resistance mechanisms for improved prognosis in difficult-to-treat tumors. Here, we describe a typical timeline of cancer resistance development, and how the technology can be used to identify signs of resistance even before treatment is given.
There are several factors that can determine whether a tumor will develop drug resistance. Cells are heterogeneous, meaning that different tumor cells can present with distinct phenotypic and morphological profiles. The novel platform has proven that cells within one patient may perform many different functions, making the use of single-cell functional measures critical to understanding patient response.
Factors such as the initial size and mutation rate of a tumor are associated with drug resistance, and changes in size, genome, and tumor microenvironment can also drive drug resistance in tumors. When a tumor responds to therapy for a period of time and then regrows, resistance may be driven by mechanisms such as new mutations, pathway alterations, or histological changes.
While the development of drug resistance in cancers is a major therapeutic hurdle, there are tools that researchers and clinicians can use to develop effective combination therapies by identifying the mechanisms of drug resistance before it occurs. Of the existing strategies used to mitigate the risk of drug resistance, including early detection of cancer and regular therapeutic monitoring, functional single-cell proteomics is the only technology that provides deep functional phenotyping of cancer cells.
Single-cell proteomics reveals multiple pathways to drug resistance
In a study published in Nature Communications, researchers Su et al. used IsoPlexis’ Single-Cell Intracellular Proteome solution to analyze mutant melanoma cancer cells (BRAFV600E M397) treated with BRAF inhibitor (BRAFi) over five days. This platform characterized the phosphoproteins and metabolites of single cells and measured oncogenic signaling, cell proliferation, and metabolic activity.
The researchers identified two different cell populations within the overall group of cells, which took different paths to drug resistance. The researchers were able to use this information to identify drug susceptibilities in both pathways and predicted that a combination therapy targeting multiple pathways would overcome drug resistance and yield the best response.
Indeed, while monotherapy or dual-inhibition approaches slowed tumor growth somewhat, the combination therapy predicted using IsoPlexis’ platform—a BRAF, NFκB, and PKM2 triple-inhibition approach—resulted in a superior response compared to any of the other drug combinations.
In their paper, Su et al. stated that IsoPlexis’ intracellular proteome and metabolome technology “update[s] the current understanding of resistance development and can provide a powerful methodology for identifying therapy combinations,” enabling researchers to develop combination therapies to combat drug-resistant cell states.

