Rapid Antimicrobial Susceptibility Testing: The Diagnostic Answer to the Superbug Crisis
Next-generation phenotyping holds promise for antibiotic de-escalation
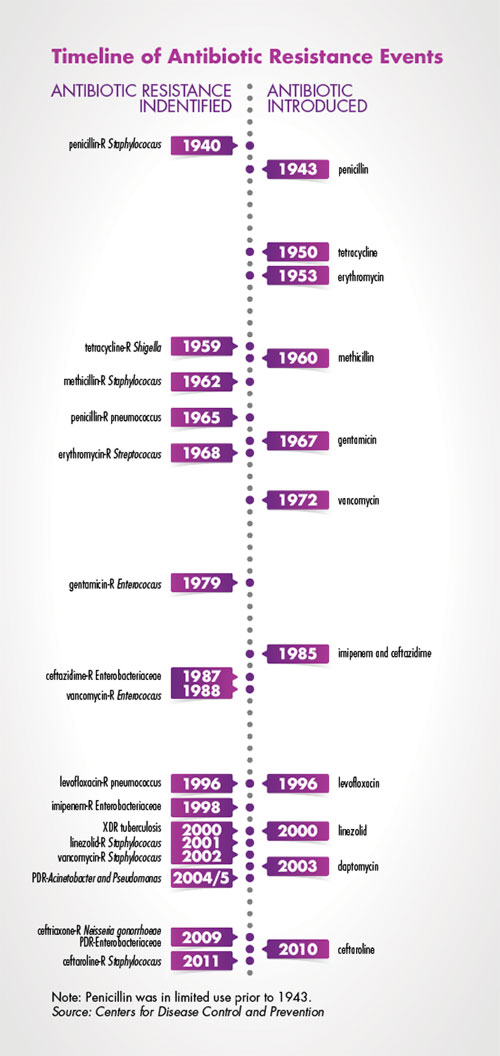
One of the most significant factors driving the antibiotic resistance crisis is the widespread use, and overuse, of broad-spectrum antibiotics. The more frequently antibiotics are deployed, the more likely it is that pathogenic microbes will evolve resistance. Under the current paradigm of clinical care, septic patients are prescribed broad-spectrum antibiotics for a minimum of three days—and often five or more days. This over-reliance on some of the best drugs currently available directly fuels the rise of multidrug-resistant organisms (MDROs).
To combat the rise of MDROs, patients must be transitioned to personalized, targeted therapies as quickly as possible. Recent investments in rapid bacterial identification (ID) and resistance marker platforms have aided clinical microbiology laboratories in their quest to help clinicians better target therapies. While these platforms rapidly differentiate viral from bacterial infections, doctors will prescribe a targeted antibiotic therapy only if they can be certain that the drug will effectively target a patient’s infecting pathogen. In the absence of rapid antimicrobial susceptibility testing (AST) platforms, infectious disease doctors will continue to over-rely on broad-spectrum therapies.
A new prescription paradigm
The era of personalized medicine effectively originated in infectious disease departments, thanks to microbial ID and AST serving as the companion diagnostics that directed therapies. The first automated ID/AST platforms enabled virtually all clinical microbiology laboratories to provide consistent, reliable results to infectious disease doctors.
While revolutionary in their time, these automated platforms now fail to meet clinical needs in two critical dimensions. First, results arrive too slowly, and sample preparation requires too many days to keep pace with cutting-edge ID techniques. Second, antibiotic menus are limited, which prevents most hospitals from having panels matching their formularies and means that patients infected with MDROs require additional reflex testing. Limited test menus are particularly insidious because the patients with the most difficult-to-treat infections end up receiving the most delayed, least effective care. A new diagnostic paradigm is therefore needed to improve patient care and maintain antibiotic potencies and efficacies.
The ID-to-AST gap
Over the past decade, advances in genetic and proteomic techniques have created a new gold standard for clinical microbiology laboratory ID. These platforms are more accurate and faster than traditional biochemical methods, which has resulted in a new clinical microbiology laboratory paradigm where ID results are known at least one to two days before AST results are available. This ID-to-AST gap accentuates the slow pace of AST and creates an agonizing delay for doctors who need the AST results in order to select the safest and most effective treatment for their patients.
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS, or MALDI) is not only better and faster, but also—depending on laboratory volumes—sometimes cheaper than traditional biochemical ID methods. As a result, nearly two-thirds of the clinical laboratories in the US have already adopted MALDI as their primary ID platform. These laboratories’ original ID/AST platforms now perform only half their originally intended service, as their biochemical ID approaches are obsolete.
For all its considerable strengths, MALDI suffers one major weakness: it requires isolated microorganism colonies because it cannot consistently detect more than one species at a time. This inability to reliably differentiate polymicrobial from monomicrobial samples is a result of the core physics of matrix ionization coupled with the time-of-flight detector. Thus, laboratories cannot rely on MALDI IDs from positive blood cultures to de-escalate therapies.
Genetic multiplex (gen-plex) technologies, both PCR- and hybridization-based, have found a powerful niche providing rapid ID from positive blood samples as well as directly from some patient samples, such as nasal swabs and respiratory and stool samples. Like MALDI, gen-plex platforms have dramatically impacted clinical microbiology laboratories. Using PCR, Cepheid set a new infection control standard by rapidly differentiating methicillin-resistant Staphylococcus aureus (MRSA) from methicillin-susceptible S. aureus (MSSA) infections from nasal swab samples. BioFire® (acquired by bioMérieux) then significantly expanded PCR’s multiplexing capabilities with the FilmArray® platform. Nanosphere’s VERIGENE® platform (acquired by Luminex) offers similar performance from positive blood cultures with its hybridization-based approach.
Since gen-plex platforms do not require isolated colonies, they typically provide IDs one day faster than MALDI does, exacerbating the ID-to-AST gap. But gen-plex speed is expensive: per-test prices are 10 to 25 times higher than those of MALDI. Nonetheless, clinical microbiology laboratories have rapidly adopted these platforms because they improve patient care and decrease lengths of stay.
In addition to providing IDs, gen-plex platforms provide information on known resistance genes in their multiplex assays. Because thousands of documented mutations confer resistance—even the utility of mecA to exclusively determine MRSA vs. MSSA is now under question—these fast antibiotic resistance tests are limited to providing information about which antibiotics will not work. Thus, resistance tests prove useful if there happens to be a known marker that requires therapy escalation, but they provide limited actionable de-escalation information.
Next-generation phenotyping (NGP) challenges
The rapid ID platforms have exacerbated the slow speed of AST, and infectious disease doctors are clamoring for earlier susceptibility results. To change infectious disease patient care, an AST platform must offer 1) a rapid time to results and 2) a comprehensive, expandable antibiotic menu. Furthermore, it is essential that such an NGP platform meets the throughput requirements of laboratories and the cost structures of infectious disease care.
The key technological challenge of rapid AST is performing accurate microorganism quantification while accounting for antimicrobial-induced morphological changes. This is particularly critical for beta-lactam agents, which account for approximately two-thirds of US antibiotic prescriptions. Bacteria susceptible to betalactams often filament or swell (forming spheroplasts or protoplasts) before lysing at antibiotic concentrations around the minimum inhibitory concentration (MIC).
Because they utilize bulk optical density (OD) measurements to assess microorganism growth, traditional, firstgeneration automated AST platforms cannot differentiate filamented or swelled susceptible cells from truly replicating resistant cells. Since filament, spheroplast, or protoplast lysis requires 7.5 hours for the fastest-replicating strains, traditional platforms are limited by the speed with which they can make accurate AST calls. This limitation leads many clinical laboratories to perform AST overnight, since standard clinical laboratory shifts are eight to 10 hours and AST results must typically be reviewed and interpreted by first-shift personnel before being reported.
In order to close the ID-to-AST gap, an NGP platform must therefore be capable of determining bacterial morphologies. The need for same-shift AST is further driven by the increasing shortage of trained medical technologists and the difficulty of filling second and third shifts—another harsh reality faced by many laboratory directors.
The menu limitation of first-generation AST platforms derives from their measurement paradigms. To provide AST results as early as possible, these machines take OD measurements of every reservoir on every card/plate every 15-20 minutes. These are combined to create growth curves, which are assessed by computer algorithms to determine MICs.
Though these platforms are powerful in concept, their reliance on growth curves limits the number of drugs that can be tested in parallel and creates system tradeoffs that decrease result accuracies. The need for repeated measurements imposes a significant engineering constraint. The time required to take each measurement and shuttle cards/plates from the incubator to the optical reader is compounded by the fact that the machines hold ~100 cards/plates at a time. Thus, the addition of a new test reservoir must come at the cost of either throughput or accuracy. Since no lab can afford to compromise throughput or accuracy, the number of test reservoirs is locked for each platform.
As a result, directors are faced with a zero-sum game when new drugs gain FDA approval: to accommodate a new drug, they must sacrifice a generic mainstay already on the panel. Adding meropenem-vaborbactam, for example, requires the sacrifice of piperacillin-tazobactam. Because Kirby-Bauer diffusion disks suffer no such trade-off, these disks are available shortly after drugs gain FDA approval.
The limit on the number of test wells on traditional AST platforms also prevents laboratories from performing actual quality control (QC) tests for many drugs. This has created such significant accuracy issues that the FDA no longer grants clearances for agents that do not include on-scale QC. Thanks to this new FDA requirement, all newly cleared drug-bug combinations on traditional AST platforms, as well as all drug-bug combinations on new AST platforms, must provide on-scale QC.
The NGP hunt continues
The hunt continues for a rapid, broad-menu, cost-effective NGP platform that brings AST into the 21st century. Results should be available within five to six hours to enable the same-shift reporting necessary to keep pace with rapid ID technologies and eliminate the ID-to-AST gap.
Furthermore, the distribution of the time to results should be minimal in order to allow hospitals to establish structured workflows for antibiotic stewardship team reporting. In addition, the system should not only provide results from a broad menu of antibiotics but also should have the capability to easily accommodate new drugs as they are developed. While meeting these requirements, the new platform must also remain more affordable than rapid gen-plex ID systems.
NGP PLATFORM WISH LIST |
|---|
The ideal NGP platform for AST should:
|
Though companion diagnostics began with AST, current AST platforms’ slow speeds and incomplete results have considerably dulled AST’s sheen. Since susceptibility results are more important than ever due to the rise of MDROs, companies and investigators are now trying to find ways to emulate phenotypic results with genetic technologies. These approaches, such as next-generation sequencing, face considerable technical and cost challenges but must be considered real competitors to NGP.
Multiple companies and investigators are now racing to develop AST platforms based on new detection paradigms. The winning NGP technologies will solve the AST speed and menu issues cost-effectively while addressing the medical technologist shortage plaguing clinical microbiology laboratories.
The need has never been greater; time will tell whether the current generation of diagnostic developers can create an NGP platform that allows AST to reclaim its companion diagnostic throne.
